Molecular Biological Analyses
1. Detection of Periodontal Bacteria
– The bacterial flora in human dental plaque is diverse and complex, consisting of more than 700 bacterial species, whereas a small subset of the species is considered to be the primary agents of periodontal diseases.
– The periodontal pathogens generally belong to the obligate anaerobe group, and isolation of these species is technically very demanding and time-consuming.
– On the other hand, the recent development of molecular biological techniques enables the analysis of the periodontitis-related species without bacterial isolation.
– The PCR technique using the bacterial DNA extracted from the specimens can identify the bacteria with species-specific sets of primers, whereas the disadvantage of this approach is that only selected species can be analyzed.
– In contrast, broad-range PCR and sequence methods can demonstrate the bacterial profiles
in the specimens since the primers are designed for the common region of the 16S rRNA sequences for eubacterial species.
– The advantage of this method is that it can identify any species registered in the genomic
data bases but it takes more time and is relatively expensive compared to the conventional PCR method.
– In addition, a quantitative PCR method can be used to determine the bacterial levels in the specimens.
– Although it would be valuable to determine the numbers of each species in the specimens, it would be relatively expensive to do so at present with large numbers of specimens in
large-scale clinical studies.
– It is also important to decide whether saliva or dental plaque specimens are to be analyzed when using these techniques.
– Whole saliva specimens generally reflect the entire bacterial profiles in the oral cavity, whereas the profiles of the dental plaque specimens are limited to those of the collected area.
– Thus, we must carefully consider which specimens match the purpose of studies prior to their initiation.
– In addition, the timing of collection of the specimens is also important. We generally collect the specimens for the initial stage at the second visit to our clinic, prior to giving detailed tooth brushing instructions, which enable us to determine the natural bacterial profiles of each subject.
– The procedures for extraction of the bacterial DNA are as follows.
– The subgingival dental plaque specimens are collected in sterile saline, which are centrifuged at 15000 rpm for 5 min to pellet the bacterial cells. Bacterial genomic DNA is extracted from each pellet using a DNA isolation kit (Puregene, Gentra Systems, Minneapolis, MN, USA).
– As for the saliva samples, expectorated whole saliva collected from each patient is mixed with Chelex 100 (Bio-Rad Laboratories, Hercules, CA, USA), which is then incubated at 56°C for 30 min, followed by boiling at 100°C for 10 min.
– Each sample is then centrifuged at 15000 rpm for 20 min and the supernatants used as
templates for PCR assays.
– Primer list for detection of 10 periodontitis-related species
Species | Sequence (5‘to 3‘) | Amplification size (bp) |
Positive control | AGA GTT TGA TCM TGG CTC AG CTG CTG CSY CCC GTA G | 315 |
Pg | TGT AGA TGA CTG ATG GTG AAA ACC ACG TCA TCC CCA CCT TCC TC | 197 |
Td | AAG GCG GTA GAG CCG CCG CTC A AGC CGC TGT CGA AAA GCC CA | 311 |
Tf | GCG TAT GTA ACC TGC CCG CA TCG TTC AGT GTC AGT TAT ACC T | 641 |
Co | AGA GTT TGA TCC TGG CTC AG GAT GCC GTC CCT ATA TAC CAT TAG G | 185 |
Cs | AGA GTT TGA TCC TGG CTC AG GAT GCC GTC CCT ATA TAC GGG G | 185 |
Pi | TTT GTT GGG GAG TAA AGC GGG TCA ACA TCT CTG TAT CCT GCG T | 575 |
Pn | ATG AAA CAA AGG TTT TCC GGT AAG CCC ACG TCT CTG TGG GCT GCG A | 804 |
Aa | AGA GTT TGA TCC TGG CTC AG CAC TTA AAG GTC CGC CTA CGT GCC | 593 |
Cr | TTT CGG AGC GTA AAC TCC TTT TC TTT CTG CAA GCA GAC ACT CTT | 598 |
Ec | CTA ATA CCG CAT ACG TCC TAA G CTA CTA AGC AAT CAA GTT GCC C | 688 |
– Various bacterial species have been reported to be related to periodontitis, among which we recently focused on 10 species, including Porphyromonas gingivalis (Pg), Tannerella
forsythia (formerly Tannerella forsythensis) (Tf), Prevotella intermedia (Pi), Prevotella
nigrescens (Pn), Campylobacter rectus (Cr), Eikenella corrodens (Ec), Aggregatibacter
(formerly Actinobacillus) actinomycetemcomitans (Aa), Capnocytophaga ochracea (Co), Capnocytophaga sputigena (Cs) and Treponema denticola (Td), based on previous reports showing that their distributions in periodontitis patients were significantly different from those in periodontally healthy subjects.
– Previous table lists the primer sets specific for each species used in our studies, all of which were checked for specificity as well as sensitivity and was reported to range from 10-100 cells in the original studies.
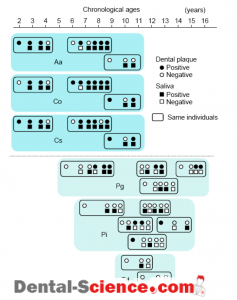
– There were two species identified in the saliva specimen of subject A, whereas 6 species were detected in the specimen taken from subject B.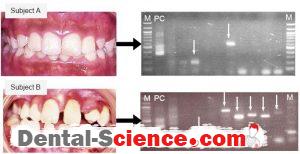
2. Distribution of Periodontal Bacteria in Children
– There have been few reports describing the results of the distribution of periodontal bacterial species in children and adolescents when we initiated this study.
– It was generally known that children with periodontitis are rarely encountered, which might be one of the reasons for the few studies analyzing their distribution.
– Therefore, we decided to analyze the distribution of the 10 selected bacterial species in the dental plaque and saliva specimens in children and adolescents who came to our clinic.
– Dental plaque specimens were collected from the buccal-mesial sulcus of the first molar or second primary molar in the right upper quadrant of 119 systemically healthy children (56 boys and 63 girls) aged 2 to 13 years old, who showed negligible periodontal inflammation, and their whole expectorated saliva specimens were also collected.
– The total numbers of dental plaque and saliva specimens collected from these subjects were 300 and 208, respectively.
– Those subjects had received no antibiotic medication for at least 3 months prior to each
specimen collection.
– PCR detection using 10 species-specific primer sets showed that approximately 15% of the subjects were negative for any of those 10 species.
– The total numbers of the detected species increased gradually with age until 5 years old and then reached a plateau after the mixed dentition period.
– The detection rates for most of the species in saliva specimens were significantly higher than those in dental plaque specimens. C. rectus, E. corrodens, A. all age groups,
while T. forsythia and P. intermedia were detected less frequently, and P. gingivalis and
T. denticola were rarely identified.
– These results suggest that the colonization of many periodontal bacterial species occurs quite early in childhood without clinical signs of periodontal disease. In contrast, there are several species exhibiting poor colonization, such as P. gingivalis and T. denticola.
– Next, in order to determine which species are inhabitants or are transient, the subjects whose specimens were collected more than twice for more than 2 years were analyzed.
– C. ochracea, C. sputigena, A. actinomycetemcomitans, P. nigrescens, C. rectus, and
E. corrodens were frequently detected in multiple specimens taken at different times,
suggesting that these are the common members of the oral flora of periodontally healthy children.
– In contrast, P. gingivalis, T. denticola, and P. intermedia were rarely detected which indicates that these species should be regarded as transient species in children. These results suggest that there exist some species with early colonization and that others are just transient
species in children.
3. Longitudinal Studies of Periodontitis-Assocated Bacteria
– It is of interest to determine the distribution of the periodontal species in subjects over time.
– The occurrence of periodontal bacterial species in 192 systemically healthy subjects (89 male and 103 females, 2–16 years old) was analyzed in 1999-2000, among which 26 subjects
continued to attend annual recall examinations until 2006–2007.
– Thus, a total of 26 children and adolescents from whom dental plaque and saliva specimens were obtained during both the 1st (1999-2000) and 2nd (2006-2007) periods, were
analyzed.
– The periodontal condition of most of the subjects in the 2nd period was relatively good, which reflected their continuous participation in our recall system, in which not only
periodical examinations were performed but also professional tooth cleaning as well as brushing instructions were provided.
– On the other hand, there were several subjects with periodontal pocket depths of 4-5 mm without attachment loss.
– The PCR method specified the presence of 10 periodontal bacterial species, which indicated
a positive correlation of periodontal pocket depth with the total number of detected
species. P. gingivalis, T. denticola, and T. forsythiacomprise the “red complex” (RC) group of species, a prototypical polybacterial pathogenic consortium active in periodontitis.
– RC species are also known to be correlated with gingival inflammation based on studies of Japanese teenagers.
– We further analyzed the bacterial profiles in the first and second collections focusing on the presence of RC species.
– Subjects with RC species in saliva specimens obtained during the 2nd collection possessed
a significantly higher number of total bacterial species than those without these organisms.
– The main reason for this result may be because P. nigrescens, C. rectus, and E. corrodens were detected at significantly higher rates in the subjects with RC species.
– The detection rate of the RC species in the 2nd collection period specimens were significantly greater in subjects who had 2 or more species detected in specimens taken during the 1st collection compared with the other subjects.
– In addition, the total number of bacterial species in each of the saliva specimens positive for RC species was greater than 4.
– Furthermore, retrospective analyses of specimens from subjects with RC species in
1999- 2000 showed that those subjects possessed a significantly greater number of total species than the subjects without RC species. When all specimens obtained at the 1st
collection were divided into 2 categories based on the number of total bacterial species, high (2 or more) and low (1 or 0), the detection rate of the RC species at the 2nd collection was significantly greater in the high group (Odds Ratio; 17.5, 95% confidence interval;
1.2-250.4).
– These results indicate that those subjects who harbor the RC species may be at possible risk for colonization by high levels of periodontal bacterial species during adolescence and early adulthood.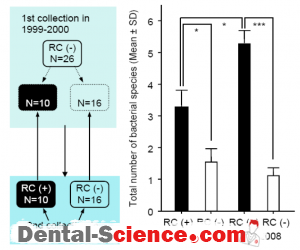
4. Multiplicity of Species Detected
– between bacterial species that reside in the biofilms are reported to influence the
composition of the communities.
– These interspecies interactions are known to play an essential role in balancing competition and coexistence.
– Furthermore, synergistic interactions may stimulate the growth or survival of one or more of the residents.
– In order to determine which species are present simultaneously in the oral cavity, we
analyzed the saliva specimens from 113 children (61 boys and 52 girls) aged 2-12 years old [64], which revealed 9 combinations of the species simultaneously detected.
– It should be noted that C. rectus and E. corrodens tended to be detected simultaneously with the RC species.
– Another study analyzing 74 children (39 boys and 35 girls) aged 2-13 years old demonstrated that the presence of C. rectus was correlated with at least one of the RC species (Odds Ratio; 10.4, 95% confidence interval; 1.8-59.6).
– The prevalence of T. forsythia and C. rectus in children was low when compared with the other species, while statistical analysis revealed that T. forsythia and C. rectus tended to be detected simultaneously in the children.
– It is of interest that the number of detected species in the children with T. forsythia and/or C. rectus was 3.71 ± 1.71, which was significantly higher than that for those without either of those species (1.15 ± 0.92).
– In addition, two children with P. gingivalis were found to possess both T. forsythia and
C.rectus.
– The other study analyzing 107 Japanese children aged 8-11 years old and adolescents aged 15 years old showed that P. gingivalis and T. forsythia, as well as T. forsythia and C. rectus were markers for increased risk of developing periodontitis.
– Taken together, it is possible to speculate that detection of these species in Japanese children and adolescents might indicate that these subjects are at high risk of developing
periodontitis in the future.
– It will be of interest to determine whether this hypothesis is applicable to other ethnic groups as well.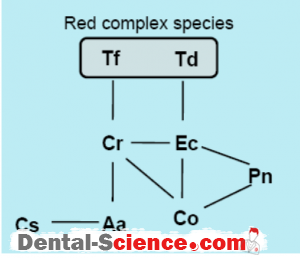
5. Mother-To-Child Transmission
– It is generally believed that Streptococcus mutans, a pathogen of dental caries, can be
transmitted from mothers to their children, and our molecular biological analyses showed that the transmission rates of S. mutans in Japanese children aged 2-10 years old was
approximately 70%.
– Another study analyzing Japanese children aged 0-11 years old showed that the transmission rate was 76.5%.
– Thus, S. mutans is frequently transmitted early from mothers to their children, which reflects an environment involving close contact between children with their mothers.
– As for the periodontal bacterial species, intra familial transmission of P. gingivalis,
A. actinomycetemcomitans, P. intermedia, and P. nigrescens, has been studied using
various methods, all of which required isolation of each strain.
– The difficulty in carrying out such approaches is the major reason why there have been only
a limited number of studies regarding mother-child transmission of periodontal bacterial species.
– However, the acquisition of periodontal organisms as the initial important step in the
development of periodontal diseases in children and adolescents has been discussed,
with a 54 Nakano, At suo Amano and Takashi Ooshima focus on intra familial transmission.
– On the other hand, PCR methods using the bacterial DNA extracted from the oral specimens can readily identify organisms in children and their mothers.
– Thus, we decided to compare the distribution of oral bacteria in children and their mothers in order to understand which periodontal bacterial species are transmitted from mothers to their children.
– Saliva specimens were collected from 113 pairs of children (61 boys and 52 girls) aged 2-12 years and their mothers aged 24-49 years.
– PCR detection identifies the species estimated to be involved in mother-to-child transmission based on the results of the detection rates in both groups. T. denticola was detected most frequently (83%) in children whose mothers possessed the same pathogen and was shown to be significantly higher than that in children whose mothers did not harbor the
spirochete.In addition to T. denticola,C. sputigena, A. actinomycetemcomitans, and
E. corrodens showed this same tendency.
– These results suggest that a correlation between the presence of periodontal bacteria in
children and their mothers, and the presence of RC bacteria in children was shown to be highly associated with the prevalence in their mothers.
– As for the other species, the detection rates for C. sputigena and A. actinomycetemcomitans were relatively high in both mothers and children.
– Thus, the presence of these species in the mother-child pairs might have been coincidental and were not statistically analyzed.
– In addition, the numbers of children with P. gingivalis were too low in the present study to estimate transmission from mother to child. However, another study demonstrated
a tendency for detection of P. gingivalis in children whose mothers were also positive for this organism.
– In addition, the genotypes of P. gingivalis fimbriae genes for mother-child pairs were shown
to be consistent which led us to speculate that P. gingivalis is also one of the species
transmitted from mothers to children.
– An additional study analyzing 56 Japanese children and adolescents aged 1-15 years and their parents suggested that P.
– As described above, RC species are reportedly associated with gingival inflammation based on studies of Japanese teenagers.
– In our study, approximately 40% of the saliva specimens taken from mothers were shown to possess RC species and their total number of periodontal bacteria was significantly higher than in those without RC species.
– The detection rate of the RC species in children whose mothers possessed the same species was shown to be significantly higher than in those whose mothers did not (odds ratio 7.4).
– A similar study conducted in the United States demonstrated that children whose parents were colonized by RC species were 9.8 times more likely to be colonized by the same
species.
– In addition, the total number of bacterial species in children whose mothers possessed the RC species was shown to be significantly greater than in those whose mothers did not.
– Taken together, the present data suggested that mothers should be careful not only
regarding the current periodontal condition of their children but also their own periodontal health in order to prevent the early subsequent onset of periodontitis in their children.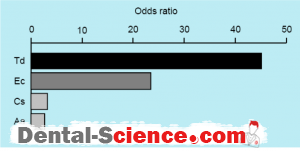
6. Porphyromonas gingivalis Fimbriae
– P. gingivalis is considered to be one of the most important pathogens in periodontal disease.
– This pathogen expresses fimbriae, filamentous appendages on the bacterial surface, which are thought to play a significant role in colonization and invasion of periodontal tissues.
– FimA, a subunit protein of the major fimbriae, is encoded by the fim A gene which is classified into six variants (types I through V and Ib).
– Net table lists the primer sets for identification of fimA genotypes in the clinical specimens.
– A majority of the P. gingivalis organisms isolated from marginal periodontitis patients are
reported to belong to type II, followed by type IV, while type I tends to be prevalent in
periodontally healthy adults.
– In fact, several other studies conducted in different countries also support our findings that type II fimA organisms are strongly correlated with the development of periodontitis.
– These findings suggest the existence of disease-associated and non-associated P. gingivalis.
– Primer list for determination of fimA genotypes of P. gingivalis
FimA genotype | Sequence (5‘to 3‘) | Amplification size (bp) |
Type I | CTG TGT GTT TAT GGC AAA CTT C AAC CCC GCT CCC TGT ATT CCG A | 392 |
Type II | ACA ACT ATA CTT ATG ACA ATG G AAC CCC GCT CCC TGT ATT CCG A | 257 |
Type III | ATT ACA CCT ACA CAG GTG AGG C AAC CCC GCT CCC TGT ATT CCG A | 247 |
Type IV | CTA TTC AGG TGC ATA TAC CCA A AAC CCC GCT CCC TGT ATT CCG A | 251 |
Type V | AAC AAC AGT CTC CTT GAC AGT G TAT TGG GGG TCG AAC GTT ACT GTC | 462 |
Type Ib | CAG CAG AGC CAA AAA CAA TCG TGT CAG ATA ATT ATT AGC GTC TGC | 294 |
– Further, 70% of periodontally healthy adults with P. gingivalis were found to carry type I, the remaining carried type V fimbriae and there were a few subjects positive for types II, IV and Ib.
– Type II was the highest, followed by types IV and Ib, all of which are regarded as highly
virulent groups, whereas types I, III and V are considered to be low virulence groups.
– In addition, the results of in vitro analyses using gingival epithelial cells as well as a mouse model of abscess formation supported this suggestion.
– Specifically, it was interesting to observe that a mutant with a substitution of the type I fimA gene with that of type II showed enhanced bacterial adhesion/invasion to epithelial cells, whereas that with substitution of type II fimA with type I resulted in diminished
adhesion/invasion.
– These results suggest that it is possible to estimate the risk of subjects for periodontitis by
analyzing the types of P. gingivalis fimA genes in oral specimens.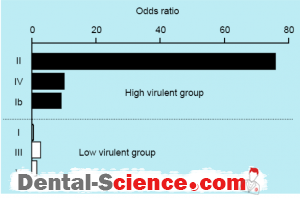
– A total of 650 saliva specimens were isolated from 464 children (3 to 18 years of age) were analyzed, and PCR detection showed that only 15 (3.23%) subjects were P. gingivalis
positive.
– The detection ratesforP. gingivalis and the ages of the subjects were similar to those in
a previous study which supports the suggestion that P. gingivalis tended to be more
frequently detected in older subjects.
– It should be noted that P. gingivalis is regarded as a transient species in children and
adolescents and the detection rates for P. gingivalisat different times was shown to be
25-67% in P. gingivalis-positive subjects.
– Therefore, multiple specimens at different times are required to identifyP. gingivalis-positive subjects.
– Furthermore, additional assays of these specimens to discriminate between the fimA
genotypes demonstrated that none of these showed a positive reaction to the type II
fimA specific primers, while 4, 1, and 2 subjects were shown to be positive for the type I, Ib, and III genotypes, respectively.
– In addition, the type IV genotype was detected in three subjects in the older age group.
– It should be noted that one-third of the fimA genotypes were determined to be un typeable in P. gingivalis-positive subjects.
– It remains to be elucidated whether the un typeable specimens contained only a single or multiple unknown fimA genotypes.
– However, it is possible that this/these genotypes possibly belong to low virulence groups, such as I, III and V. Taken together, these results suggest that the distribution of type II and IV fimA genotypes is extremely low in children although only a limited number of children do harbor P. gingivalis with low virulence for periodontitis.
– Furthermore, some adolescents were found to possess the type IV fimA genotype, which are possibly related to the onset of marginal periodontitis.
– un typeable specimens contained only a single or multiple unknown fimA genotypes.
– However, it is possible that this/these genotypes possibly belong to low virulence groups, such as I, III and V. Taken together, these results suggest that the distribution of type II and IV fimA genotypes is extremely low in children although only a limited number of children do harbor P. gingivalis with low virulence for periodontitis.
– Furthermore, some adolescents were found to possess the type IV fimA genotype, which are possibly related to the onset of marginal periodontitis.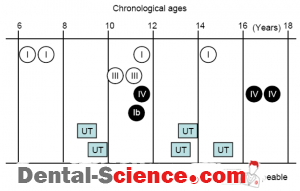
7. Down’s Syndrome Subjects
– Down ‘s syndrome (DS) is known to be a genetic disease resulting from trisomy of the 21st chromosome, occurring in 1 out of 600-1000 births.
– It is widely known that subjects with DS often develop extensive gingivitis at early stages and exhibit extensive rapid and generalized periodontal breakdown in early adulthood, which is estimated to result from impaired immune responses, fragile periodontal tissue and early senescence.
– Subjects with DS are commonly encountered in daily dental practice; however, there is little information on periodontal bacterial species using molecular biological techniques in these subjects.
– Thus, we decided to analyze the bacterial species in DS subjects.
– The distribution of periodontal species in 60 children with Down ‘s syndrome (2 to 13 years old, 5 in each age bracket) was compared with those of 60 age-matched systemically healthy controls.
– There were no obvious signs of marginal periodontitis in the DS group and no significant
clinical difference from the control group.
– PCR detection of the 10 periodontitis-related species in subgingival plaque specimens showed that most of the pathogens were detected with greater frequency in the DS children than in the controls.
– T. denticola, T. forsythia, P. nigrescens, and C. rectus were significantly prevalent throughout all age brackets of the DS children.
– The occurrence of P. gingivalis was also significant in the DS subjects over 5 years old.
– These results demonstrated that important pathogens for several types of adult periodontitis, such as P. gingivalis, T. forsythia and T. denticola, are frequently found in DS groups.
– Although these species are considered to be transient in systemically healthy children, early colonization could occur in the DS group.
– In addition, the severity of gingivitis was associated with increased varieties of the resident pathogens as well as the distribution of P. gingivalis.
– Analyses of subgingival plaque specimens from DS patients also showed that early-onset
periodontitis in DS is mainly due to the increased susceptibility of the host to the causative microbial agents including
P. gingivalis with type II fim Agenes.
– Furthermore, it was demonstrated that gingival fibroblasts from subjects with DS were
impaired significantly by P. gingivalis type II FimA as compared to those from systemically healthy subjects.
– This impairment is likely due to invasion of P. gingivalis readily leading to impaired cellular motility, which is estimated to prevent wound healing and the regeneration of periodontal tissues.
8. Children with Developmental Disabilities
– Maintenance of good oral hygiene is generally considered to be difficult in subjects with
developmental disabilities.
– This leads to speculation that the distribution of periodontal species is different in these
subjects as compared to systemically healthy individuals.
– However, there are few reports describing these species in children with developmental
disabilities.
– Therefore, we determined the distribution of the periodontal bacterial species in subjects
attending daycare centers due to the developmental disabilities.
– A total of 187 children (136 boys, 51 girls) aged 1-6 years attending daycare centers, were
analyzed.
– They were diagnosed with mental retardation (MR), cerebral palsy (CP), autism (AU),
or pervasive developmental disorders. Dental plaque specimens were collected from the buccal side of the maxillary left second primary molar.
– PCR analyses demonstrated that C. sputigena was the most frequently detected species (28.3%), followed by A. actinomycetemcomitans (20.9%) and C. rectus (18.2%).
E. corrodens, C. ochracea, and P. nigrescens were detected in approximately 10% of the specimens, whereas T. denticola, T. forsythia, and P. intermedia were rarely found, and P. gingivalis was not detected in any of the subjects.
– The mean value for the total number of the 10 tested bacterial species in all subjects was 1.16 species, which was positively correlated with age.
– In addition, the total numbers of detected species were positively correlated with the age of the subjects, with a maximum of 8 species identified in one subject.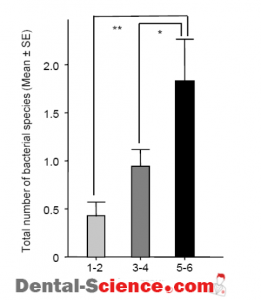
– There were 10 subjects with positive reactions for T. denticola and/or T. forsythia, in whom the total number of bacterial species was significantly higher as compared to the other
subjects.
– Furthermore, subjects possessing C. rectus showed significantly greater values for
periodontal pocket depth, gingival index, and total number of species, indicating that
C. rectus is one of the possible indicators for risk of periodontitis.
– On the other hand, the clinical parameters evaluating periodontitis were shown to be worse in the group of subjects complicated with concomitant MR, CP, and AU.
– The total number of the species detected in this group of subjects was significantly higher than those of the other groups complicated with a single or two concomitant diseases of MR, CP, and AU.
– It is reasonable to understand that individuals with multiple developmental disabilities have great difficulties with maintaining oral health and undergoing dental treatments
as compared to subjects with a single disability.
– In addition, the present results showed that one-fourth of the subjects with disabilities were shown to possess at least one of the periodontitis-associated species (T. denticola,
T. forsythia, and C. rectus) and should also be regarded as possible subjects at risk for the onset of periodontitis.
– These subjects are currently receiving special periodical professional oral health care.
– Periodical PCR detection of the species could be beneficial for evaluating their current oral status and to estimate their future risk for the onset of periodontitis.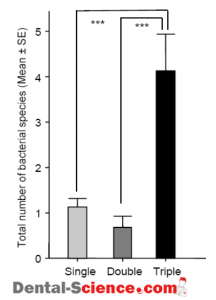
ــــــــــــــــــــ► ⒹⒺⓃⓉⒶⓁ–ⓈⒸⒾⒺⓝⓒⒺ ◄ــــــــــــــــــــ